At the Montefiore Institute (University of Liège, Belgium), we are computer science researchers developing machine/deep learning algorithms and big data software modules to make life easier for multidisciplinary teams who have to deal with very large imaging data by enabling collaboration through sharing of images, algorithms, and results over the web.
The Cytomine, open-source, software (with a permissive licence) is a rich internet application using modern web and container technologies, databases, and machine/deep learning to foster active and distributed collaboration and ease large-scale image exploitation. The software can be used remotely, using a web browser, by life scientists to help them better evaluate drug treatments or understand biological processes using various imaging modalities (including whole-slide tissue images), by pathologists to share and ease their diagnosis, by teachers and students for image-based training purposes (e.g. histology courses), and by computer scientists who want to apply their image recognition algorithms on large imaging dataset. Beyond biomedicine, the software can also being used in various other application domains that involve large image datasets such as digital collections, industrial quality control, ....
In our research team, we continously contribute to the Cytomine open-source project initiated by our research unit in 2010. The official, base, version is validated and maintained by the Cytomine cooperative company. Our Cytomine ULiège version is forked from the official version and includes base features plus experimental features driven by our research projects and collaborations, but these are not yet fully validated.
Our main research interests and contributions are:
Ongoing research & results
Here is an overview (PDF) of our research results, and illustrations below. In addition to our open-access scientific publications, our latest results are distributed through the ULiège's Cytomine open-source software repository (and later through the official repository managed by the cooperative).
-
Image segmentation techniques for the quantification of whole tissue slides.
See e.g. Marée et al. ISBI 2014. -
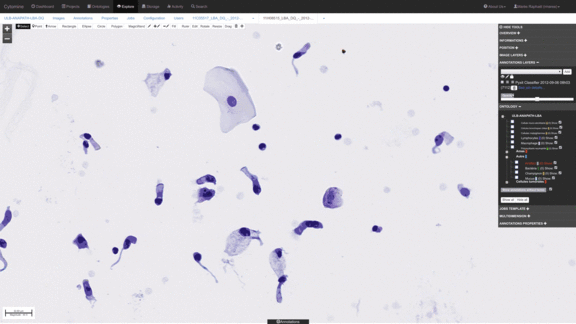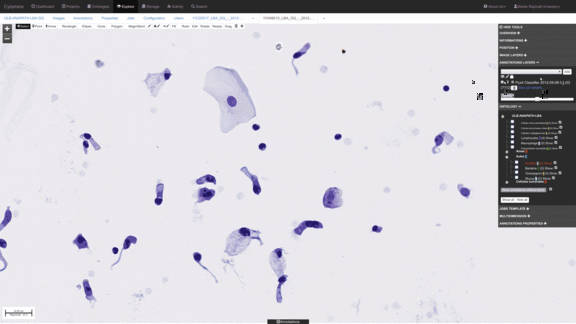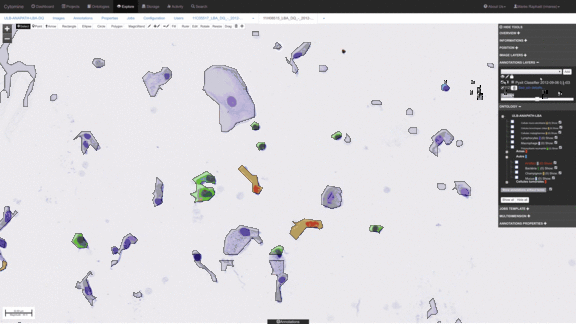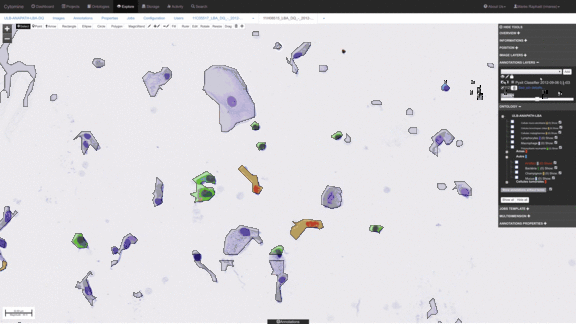
Workflows for sorting various types of cells, e.g. detect abnormal cells for early cytological diagnosis.
See e.g. Delga et al., Acta Cytologica 2014; Mormont et al., 2016. -
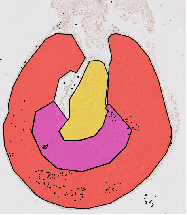
Cell counting algorithms in specific regions of interests within tissues.
See e.g. Rubens et al. (in preparation). -
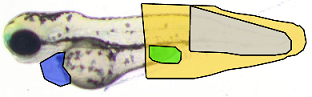
Image classification and object recognition algorithms for diagnostic of for phenotyping, e.g. in developmental and toxicological studies.
See e.g. Marée et al., PRL 2016; ISBI 2016 ; Jeanray et al., PLOSOne 2015; Mormont et al. CVPRW 2018 & IEEE JBHI 2020. -
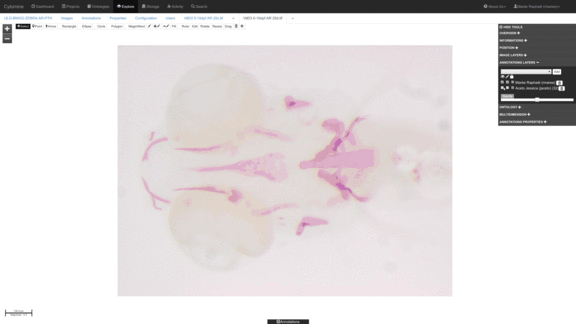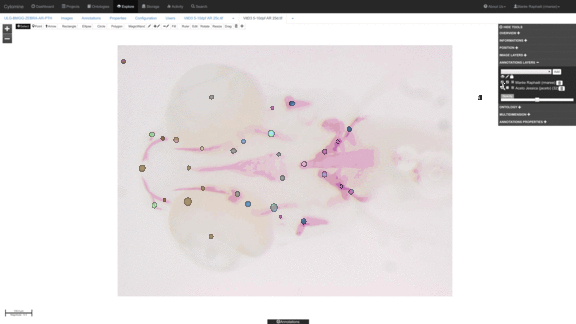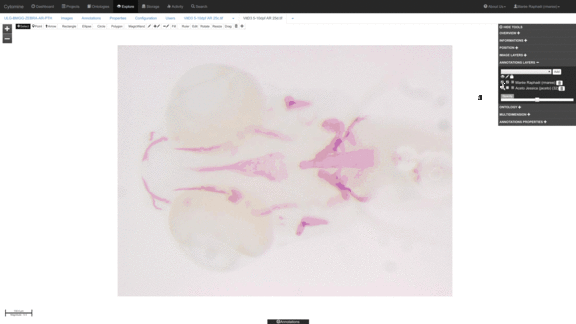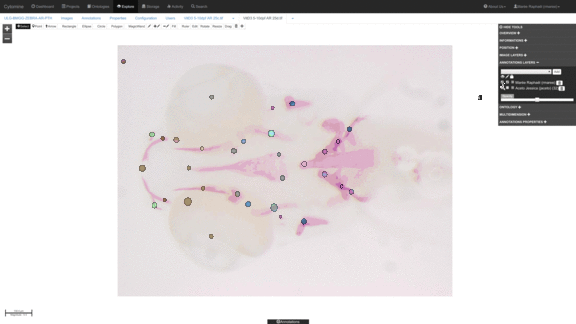
Anatomical landmark detection algorithms for morphometric change measurements e.g. in developmental and toxicological studies.
See e.g. Vandaele et al. Nature Scientific Reports, 2018. -

Tools for user behavior analytics e.g. in educational settings (Vanhee et al., Journal of Pathology Informatics, 2019).
-

User interfaces for multimodal datasets e.g. MS/multispectral imaging (Rubens et al., Proteomics Clin Appl, 2019).
-

Reproducibility and interoperability for deployment and benchmarking of image analysis workflows (Rubens et al., Cell Patterns, 2020).
-
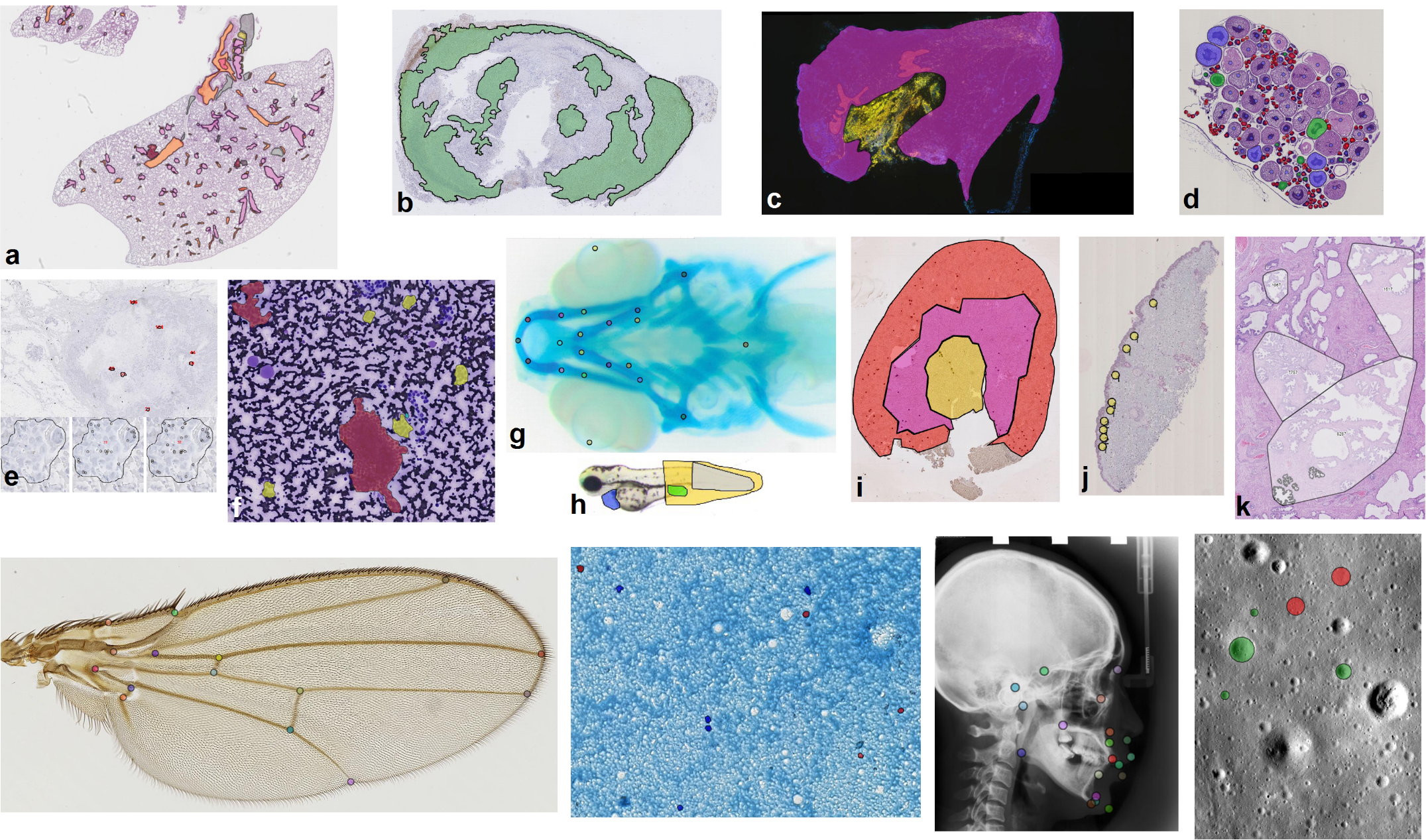
Applications in various domains (not only histology) (see publications).
News (Follow @cytomine_uliege for more regular news)
(January 2021-2027) We are involved in the BigPicture EU IMI project to establish the biggest database of pathology images to accelerate the development of artificial intelligence in medicine.
(October 2018-...) We are involved in the COMULIS COST network for correlative microscopy.
(October 2017-...) The NEUBIAS COST network WG5 choose the Cytomine software to develop its benchmarking platform (BIAFLOWS).
(1 oct. 2017) Ulysse joined our research team @ ULiège. He is now our core developer working on extensions of Cytomine for 16bits/32bits/etc. images and machine/deep learning algorithms.
(15 Sept. 2017) Grégoire, Christopher, and Renaud are now working for Cytomine.coop, a social cooperative to manage the open-source community and to provide services on top of Cytomine open-source software. Learn more about their services and their open company philosophy on Cytomine.coop.
(Sept. 2017) The first worldwide massive online course (MOOC) in french on histology is over. It was a great success ! It will come back next year. It was organized by University of Liege and the FUN platform. More info, course teaser, and registration.
(May 2016) Thanks to Regional-IT, Daily Science, and Le 15e jour for press coverage of our project and open-source release party.
(Jan. 2016) Our Cytomine paper is formally accepted for publication in Bioinformatics journal.
Cytomine is now released under an open-source software license and available on Github (see instructions below).
You can still request an account on our demo server.
(Sept. 2014) A vulgarization of some our research is available on University of Liege Reflexions website
(Oct. 2010) Cytomine kick-off.
Publications
The main Cytomine publication is (Marée et al., Bioinformatics 2016). We list here our related publications.
- Methodological papers
-
- "A rich internet application for remote visualization and collaborative annotation of digital slide images in histology and cytology"
Raphaël Marée, Benjamin Stevens, Loic Rollus, Natacha Rocks, Xavier Moles-Lopez, Isabelle Salmon, Didier Cataldo, Louis Wehenkel
BMC Diagnostic Pathology, 8(Suppl 1):S26 (Proceedings 12th European Congress on Telepathology and 5th International Congress on Virtual Microscopy), 30th September 2013 - "Extremely Randomized Trees and Random Subwindows for Image Classification, Annotation, and Retrieval"
Raphaël Marée, Louis Wehenkel, Pierre Geurts
Invited chapter in Criminisi, A; Shotton, J (Eds.) Book on Decision Forests in Computer Vision and Medical Image Analysis, Advances in Computer Vision and Pattern Recognition, February 2013 - "A hybrid human-computer approach for large-scale image-based measurements using web services and machine learning"
Raphaël Marée, Loic Rollus, Benjamin Stevens, Gilles Louppe, Olivier Caubo, Natacha Rocks, Sandrine Bekaert, Didier Cataldo, Louis Wehenkel
Proceedings IEEE International Symposium on Biomedical Imaging, pp. 902-906, 2014. - "Evaluation and Comparison of Anatomical Landmark Detection Methods for Cephalometric X-Ray Images: A Grand Challenge"
Cheng-Ta Huang, Meng-Che Hsieh, Chung-Hsing Li, Sheng-Wei Chang, Wei-Cheng Li, Remy Vandaele, Raphaël Marée, Sebastien Jodogne, Pierre Geurts, Cheng Chen, Guoyan Zheng, Chengwen Chu, Hengameh Mirzaalian, Ghassan Hamarneh, Tomaz Vrtovec and Bulat Ibragimov.
IEEE Transactions on Medical Imaging. 2015 Sep;34(9):1890-900. - "An approach for detection of glomeruli in multisite digital pathology".
Raphaël Marée, Stephane Dallongeville, Jean-Christophe Olivo-Marin, Vannary Meas-Yedid.
Proc. 13th IEEE International Symposium on Biomedical Imaging (ISBI), 2016.
- "Towards Generic Image Classification using Tree-based Learning: an Extensive Empirical Study".
Raphaël Marée, Pierre Geurts, Louis Wehenkel.
Pattern Recognition Letters, DOI: 10.1016/j.patrec.2016.01.006, 2016.
- "Collaborative analysis of multi-gigapixel imaging data using Cytomine" [Supplementary Data]
Raphaël Marée, Loïc Rollus, Benjamin Stévens, Renaud Hoyoux, Gilles Louppe, Rémy Vandaele, Jean-Michel Begon, Philipp Kainz, Pierre Geurts and Louis Wehenkel
Bioinformatics, DOI: 10.1093/bioinformatics/btw013, 2016. - "SLDC: an open-source workflow for object detection in multi-gigapixel images".
Romain Mormont, Jean-Michel Begon, Renaud Hoyoux, Raphaël Marée.
The 25th Belgian-Dutch Conference on Machine Learning (Benelearn), 2016.
- "The need for careful data collection for pattern recognition in digital pathology".
Journal of Pathology Informatics 2017, 8:19.
Raphaël Marée - Landmark detection in 2D bioimages for geometric morphometrics: a multi-resolution tree-based approach.
Rémy Vandaele, Jessica Aceto, Marc Muller, Frédérique Peronnet, Vincent Debat, Ching-Wei Wang, Cheng-Ta Huang, Sébastien Jodogne, Philippe Martinive, Pierre Geurts, and Raphaël Marée. Nature Scientific Reports, Volume 8, Article number: 5, 38 (2018). - Comparison of deep transfer learning strategies for digital pathology
Romain Mormont, Pierre Geurts, Raphaël Marée
Proc. CVPR Workshop on Computer Vision for Microscopy Image Analysis (CVMI), 2018 - Cytomine: toward an open and collaborative software platform for digital pathology bridged to molecular investigations
Ulysse Rubens, Renaud Hoyoux, Laurent Vanosmael, Mehdy Ouras, Maxime Tasset, Christopher Hamilton, Rémi Longuespée, Raphaël Marée.
Proteomics: Clinical Applications, Special Issue on Proteomics in Pathology, Volume 13, Issue 1, 2019. - New Cytomine modules for user behavior analytics in digital pathology
Laurent Vanhee, Ulysse Rubens, Alodie Weatherspoon, Laurence Pesesse, Pascale Quatresooz, Sylvie Multon, Valérie Defaweux, Pierre Geurts, Raphaël Marée
Accepted for publication in Journal of Pathology Informatics, 2019. - Open Practices and Resources for Collaborative Digital Pathology
Raphaël Marée.
Frontiers in Medicine - Pathology, Research Topic: Computational Pathology, 2019. - Multi-task pre-training of deep neural networks for digital pathology
Romain Mormont, Pierre Geurts, Raphaël Marée.
IEEE Journal of Biomedical and Health Informatics, May 2020. (arxiv version) - BIAFLOWS: A collaborative framework to reproducibly deploy and benchmark bioimage analysis workflows
Ulysse Rubens, Romain Mormont, Lassi Paavolainen, Volker Baecker, Benjamin Pavie, Leandro A. Scholz, Gino Michiels, Devrim Ünay, Graeme Ball, Renaud Hoyoux, Rémy Vandaele, Ofra Golani, Stefan G. Stanciu, Natasa Sladoje, Perrine Paul-Gilloteaux, Raphaël Marée, Sébastien Tosi.
Cell Patterns, June 2020.
- "A rich internet application for remote visualization and collaborative annotation of digital slide images in histology and cytology"
- Papers that we know using the Cytomine software or algorithm variants (non-exhaustive list, see also citations on Google Scholar)
- "Biointerface multiparametric study of intraocular lens acrylic materials"
Bertrand V, Bozukova D, Svaldo Lanero T, Huang YS, Schol D, Rosiere N, Grauwels M, Duwez AS, Jerome C, Pagnoulle C, De Pauw E, De Pauw-Gillet MC.,
J Cataract Refract Surg. 2014 Sep;40(9):1536-44. doi: 10.1016/j.jcrs.2014.01.035. - "Evaluation of CellSolutions BestPrep(R) Automated Thin-Layer Liquid-Based Cytology Papanicolaou Slide Preparation and BestCyte(R) Cell Sorter Imaging System"
Delga A, Goffin F, Marée R, Lambert C, Delvenne P.
Acta Cytologica, Vol. 58, No. 5, 2014. - "Phenotype Classification of Zebrafish Embryos by Supervised Learning"
N. Jeanray,R. Marée, B. Pruvot, O. Stern, P. Geurts, L. Wehenkel, M. Muller.
PLoS ONE 10(1): e0116989, 2015. - "Activation of the calcium-sensing receptor before renal ischemia/reperfusion exacerbates kidney injury"
Weekers L, de Tullio P, Bovy C, Poma L, Marée R, Bonvoisin C, Defraigne JO, Krzesinski JM, Jouret F.
Am J Transl Res. 2015 Jan 19;7(1):128-38 - "Zebrafish bone and general physiology are differently affected by hormones or changes in gravity"
Jessica Aceto, Rasoul Nourizadeh-Lillabadi, Raphaël Marée, Nathalie Jeanray, Louis Wehenkel, Peter Alestrom, Jack van Loon, Marc Muller.
PLoS ONE 10(6): e0126928, 2015 - "Soluble factors regulated by epithelial-mesenchymal transition mediate tumour angiogenesis and myeloid cell recruitment"
Suarez-Carmona, M. et al.
Journal of Pathology, doi: 10.1002/path.4546, 2015 - "The timing of surgery after neoadjuvant radiotherapy influences tumor dissemination in a preclinical model"
N. Leroi, NE. Sounni, E. Van Overmeire, S. Blacher, R. Marée, JA. Van Ginderachter, F. Lallemand, E. Lenaerts, PA. Coucke, A. Noel, Ph. Martinive.
Oncotarget. 2015 Nov 3;6(34):36825-37. 2015. - "Evaluation of BRCA1-related molecular features and microRNAs as prognostic factors for triple negative breast cancers"
Meriem Boukerroucha, Claire Josse, Sonia El-Guendi, Bouchra Boujemla, Pierre Freres, Raphaël Marée, Stephane Wenric, Karin Segers, Joelle Collignon, Guy Jerusalem and Vincent Bours.
BMC Cancer 16:755, 2015 - "A laser microdissection-based workflow for FFPE tissue microproteomics: important considerations for small sample processing"
Rémi Longuespée, Deborah Alberts, Charles Pottier, Nicolas Smargiasso, Gabriel Mazzucchelli, Dominique Baiwir, Mark Kriegsmann, Michael Herfs, Jörg Kriegsmann, Philippe Delvenne, Edwin De Pauw
Methods, doi:10.1016/j.ymeth.2016.12.008. - OLFM4, KNG1 and Sec24C identified by proteomics and immunohistochemistry as potential markers of early colorectal cancer stages.
Florence Quesada-Calvo, Charlotte Massot, Virginie Bertrand, Remi Longuespee, Noella Bletard, Joan Somja, Gabriel Mazzucchelli, Nicolas Smargiasso, Dominique Baiwir, Marie-Claire De Pauw-Gillet, Philippe Delvenne, Michel Malaise, Carla Coimbra Marques, Marc Polus, Edwin De Pauw, Marie-Alice Meuwis, Edouard Louis.
Clinical Proteomics, 2017. - Inflammation-Generated Extracellular Matrix Fragments Drive Lung Metastasis.
Sandrine Bekaert, Marianne Fillet, Benoit Detry, Muriel Pichavant, Raphaël Marée, Agnès Noel, Natacha Rocks, Didier Cataldo
Cancer Growth Metastasis. 2017 Dec 21. doi: 10.1177/1179064417745539 - A Massive Open Online Course (MOOC) on pratical histology: A goal, a tool, a large public ! Return on a first experience.
Sylvie Multon, Laurence Pesesse, Alodie Weatherspoon, Sandra Florquin, Jean-François Van de Poel, Pierre Martin, Grégoire Vincke, Renaud Hoyoux, Raphaël Marée, Dominique Verpoorten, Pierre Bonnet, Pascale Quatresooz, Valérie Defaweux. Annales de Pathologie, 2018. -
Basal cell carcinoma detection in full field OCT images using convolutional neural networks
D. Mandache ; E. Dalimier ; J. R. Durkin ; C. Boceara ; J.-C. Olivo-Marin ; V. Meas-Yedid
Proc. IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) -
Cell Classification in ER-Stained Whole Slide Breast Cancer Images Using Convolutional Neural Network
Jamaluddin MF, Fauzi MFA, Abas FS, Lee JTH, Khor SY, Teoh KH, Looi LM.
Proc. 40th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2018. -
Crowdsourcing of Histological Image Labeling and Object Delineation by Medical Students
A. Grote, N. Schaadt, G. Forestier, C. Wemmert, F. Feuerhake
IEEE Transactions on Medical Imaging ( Volume: 38 , Issue: 5 , May 2019) - CEMIP (KIAA1199) induces a fibrosis-like process in osteoarthritic chondrocytes
Deroyer C, Charlier E, Neuville S, et al.
Cell Death Dis. 2019;10(2):103. Published 2019 Feb 4. doi:10.1038/s41419-019-1377-8 - Ozone-primed neutrophils promote early steps of tumor cell metastasis to lungs by enhancing their NET production
Natacha Rocks, Céline Vanwinge, Coraline Radermecker, Silvia Blacher, Christine Gilles, Raphaël Marée, Alison Gillard, Brigitte Evrard, Christel Pequeux, Thomas Marichal, Agnès Noel, Didier Cataldo.
Thorax. 2019 May 29. doi: 10.1136/thoraxjnl-2018-211990. - Microproteomic Profiling of High‐Grade Squamous Intraepithelial Lesion of the Cervix: Insight into Biological Mechanisms of Dysplasia and New Potential Diagnostic Markers
Charles Pottier Mark Kriegsmann Deborah Alberts Nicolas Smargiasso Dominique Baiwir Gabriel Mazzucchelli Michael Herfs Margaux Fresnais Rita Casadonte Philippe Delvenne Edwin De Pauw Rémi Longuespée
Proteomics Clinical Applications - Special Issue: Proteomics in Pathology, Volume 13, Issue 1, January 2019 -
Strategies for Training Stain Invariant CNNs
Thomas Lampert, Odyssée Merveille, Jessica Schmitz, Germain Forestier, Friedrich Feuerhake, Cédric Wemmert
Proc. IEEE International Symposium on Biomedical Imaging, 2019
- Icytomine: A User-Friendly Tool for Integrating Workflows on Whole Slide Images
Daniel Felipe Gonzalez ObandoDiana MandacheJean-Christophe Olivo-MarinVannary Meas-Yedid
Proceedings European Conference on Digital Pathology, 2019. LNCS, vol 11435. Springer. -
Toward diagnostic relevance of the αVβ5, αVβ3, and αVβ6 integrins in OA: expression within human cartilage and spinal osteophytes
E. Charlier et al., Bone Research, 2020. -
Interobserver reproducibility of perineural invasion of prostatic adenocarcinoma in needle biopsies
Egevad et al., Virchows Archiv, 2021. - ...
- Abstracts, posters, oral presentations
- 6th Workshop on Microscopic Image Analysis with Applications in Biology (MIAAB, Heidelberg, September 2011, Poster)
- 6th IAPR Conference on Pattern Recognition in Bioinformatics (PRIB, Delft, November 2011, Poster)
- Invited talk @ 2nd European Zebrafish PI meeting (Karlsruhe, March 2012)
- Eurobioimaging workshop on Bioimage Analysis (ISBI, Barcelona, May 2012, Poster)
- 12th European Congress on Telepathology and 5th International Congress on Virtual Microscopy (Venice, June 2012)
- BioImage Informatics (Dresden, September 2012)
- Turning Images to Knowledge: Large-Scale 3D Image Annotation, Management, and Visualization (Janelia Farm, October 2012, Poster)
- Journees BioImage Informatics du GdR 2588/MIV (Paris, July 2013, Poster)
- Master Class Life Sciences Innovative Imaging of brain, body, and beyond, Hasselt University (Bioville), October, 16th, 2013
- Seminar at Centre Ingenierie Biomedicale, ULB, Brussels, October, 29th, 2013
- Liege Creative, Liege, November, 21th, 2013
- 3rd European Conference on Whole Slide Imaging and Analysis, Heidelberg, November 29 - 30, 2013
- Big Data workshop, UniGR, Luxembourg City, December 6th, 2013
- forPath Workshop on Security and Informatics in Pathology, Brussels, December 7th, 2013
- Benelux Bioinformatics Conference, Brussels, December 9th-10th, 2013 (Oral presentation prize winner)
- Workshop Intercanceropole - Microscopie Virtuelle, Paris, February 5th, 2014
- 12th European Congress on Digital Pathology, Paris, June 2014(Poster)
- BioImage Informatics, Leuven, October 2014 (Poster)
- European BioImage Analysis Symposium, January 5th-6th 2015, Paris
- GdR ISIS "Analyse de tissu biologique et histopathologie numerique", June 23rd 2015, Paris
- 14th International Congress for Stereology and Image Analysis, July 7th-10th 2015, Liege, Belgium
- Digital Pathology Asia, September 2015, Kuala Lumpur, Malaysia
- GDR MIV workshop "Bioimage Informatics", November 23-24th 2015, Paris, France.
- BigDataWeek Wallonia, November 24-25th 2015, CETIC, Charleroi, Belgium
- Journee Traitement d'images en Microscopie: "Cycle de vie des images en microscopie: les enjeux techniques", December 15th, Jussieu, Paris, France.
- FOSDEM'16 Lightning talk, January 30th 17h (H.2215 (Ferrer)), Brussels, Belgium. (Video of our talk)
- Séminaire Life Science, Universite de Namur, 3 mars 2016.
- Cytomine open source release party, 25th March 2016, Montefiore Institute, Liège.
- Med-e-Tel (International eHealth, Telemedecine and Health ICT forum), 6th April 2016 Conference Room 4 14h, Luxembourg.
- International Symposium on Biomedical Imaging (ISBI'16), 13-16th April 2016, Prague, Czech Republic.
- AIDPATH workshop, 25th April 2016, Kortrijk, Belgium. (Our slides)
- 13th European Congress on Digital Pathology (ECDP 2016), 25-28th May 2016, Berlin, Germany.
- Seminar at Image and Signal Processing Group, UCL, Louvain-La-Neuve, Belgium, 9th June 2016 (14h, "Shannon" Seminar Room, Place du Levant 3, Maxwell Building, 1st floor).
- Workshop "Bioinformatics meets digital pathology" at ECCB, 3th September 2016, Den Hague, The Netherlands.
- Pecha Kucha Open Science night, November 24th 19h, Salle Val Benoit, Liege, Belgium.
- NEUBIAS talk and open software showcase, 15-17th February, Lisbon, Portugal.
- Open source village at Med-e-Tel, 5th-7th April 2017, Luxembourg.
- France Bioimaging 4th Annual meeting, 14th April, Institut Curie, Paris, France.
- Nuit des chercheurs - Intelligence artificielle et réalité virtuelle. 29 septembre 2017 17h. Médiacité, Liège.
- Invited talk @ 4th Digital Pathology Congress, Talk on 1st Dec 2017 (2.45PM, Room Lindbergh 2&3), London, UK.
- Invited talk @ VIB Image processing workshop, Talk on 5th December (3.30PM), Leuven, Belgium.
- NEUBIAS Conference, 27 Jan - 2 Feb 2018, Szeged, Hungary.
- Oral presentation and Cytomine SCRL FS booth @ European Conference on Digital Pathology, 29th May - 1st June 2018, Helsinki, Finland.
- Invited talk @ 1ère Journée autour des mathématiques et de l’informatique en analyse de données et d’imagerie en oncologie, 12 juin 2018, Nancy.
- Demo booth @ Conference on Computer Vision and Pattern Recognition (CVPR), 20th June 2018, Salt Lake City, USA.
- Invited talk @ Medical University Graz, 20th December 2018.
- 3rd NEUBIAS Conference, Luxembourg, 6th-8th February 2019.
- Invited talk at Pathologie digitale: enjeux et opportunités, 13th March 2019, Institut d'Études Avancées de Paris (Hôtel de Lauzun), France.
- Invited talk @ 17ème édition d’ORASIS, journées francophones des jeunes chercheurs en vision par ordinateur, 30 mai 2019, Saint-Dié-des-Vosges, France.
- Colloque International "La généalogie des formes dans la perspective du Deep Learning", 7-8 novembre 2019, Liège (Belgium).
- COMULIS conference, 21st-22th November 2019, Vienna (Austria).
- Neubias Conference, 4-6th March 2020, Bordeaux (France).
- COMULIS (EU COST Action) virtual training school on "Image processing for correlated and multimodal imaging techniques", 19th October 2020, VIB.
- 3rd virtual school of the Biomedaqu ITN project, 19th November 2020.
- I2K (Images to Knowledge) Online Tutorial on BIAFLOWS, December 1st-2nd 2020.
- Meet us at upcoming events (talks/posters/showcases)
-
- ...
Software and source code
We release our research results mainly through the R&D Cytomine open-source software ULiège repository, while the official, base version is available on the official Cytomine repository.
To install and use the ULiège R&D version you can follow experimental version installation instructions and its full documentation wiki (it includes guides for administrators, developers, and a Cytomine guide for end-users). To install the official, base version, please follow official installation instructions
Cytomine can be installed on large servers for large-scale studies but also on laptops for small-scale works (then loosing collaboration functionalities). You can also check the Cytomine user guide to get access information on our R&D demo server using default accounts. Access to an official, stable, version can be requested to Cytomine cooperative.
We kindly ask you to cite Cytomine website url (www.cytomine.org) and our main publication (Marée et al., Bioinformatics 2016) when you use our software in your own work.
Research team members and collaborators
Raphaël Marée initiated the Cytomine research project in 2010. He is a senior researcher in machine learning/big data/computer vision and supervises the Cytomine big data research and development activities at ULiège.
Ulysse Rubens is a software developer (Cytomine core modules and machine learning).
Romain Mormont (PhD Student) is a researcher in machine learning (deep learning, transfer learning).
Navdeep Kumar (PhD student) is a researcher in machine learning (deep learning, domain adaptation for multimodal images).
Prof. Pierre Geurts and Prof. Louis Wehenkel are long-term machine learning collaborators.
Previous contributors (team members or students) include: Loic Rollus (2010-2015), Benjamin Stevens (2010-2014), Renaud Hoyoux (2014-2017, now at Cytomine cooperative), Gilles Louppe (2013), Jean-Michel Begon (2014), Pierre Ansen, Julien Confetti, Olivier Caubo, Thomas Vessiere, Laurent Vanosmael (2017), Gino Michiels (2018), Laurent Vanhee (2018), Mehdy Ouras (2018), Guillaume Vissers (2019), Loïc Sacré (2019), Rémy Vandaele (2014-2019),...
Through various collaborations (including the NEUBIAS COST Action), other contributors were/are coming from IRB Barcelona, Pasteur Institute (Paris), VIB Ghent, MRI Biocampus Montpellier, ...
Developments and services related to the base, official, version are lead by the Cytomine SCRLFS team.
Contact
We recommend to use the Image.sc forum as the discussion channel for user related questions.
For bugs or feature requests we recommend to post your issue on our Github repository.
For any questions related to our research activities or for potential research collaborations or internships @ ULiège, feel free to contact us by e-mail: Raphaël Marée.
To ask for a specific demo account, to support the open-source project, or for service requests (e.g. installation, hosting, maintenance, trainings, specific software developments, slide scanning), please contact Cytomine cooperative.
Finally, if you want to visit us, we are based here:
Quartier Polytech 1, 10, Allée de la découverte
Montefiore Institute (B28)
Tel. : +32 4 366 26 44
Funding
We are/were involved in the following projects:
This project will establish the biggest database of pathology images to accelerate the development of artificial intelligence in medicine.
This project aims at developing new image analysis modules for Zebrafish imaging.
This project aims at developing new visualization and annotation modules for video data.
This project aims at developing new multimodal imaging visualization and annotation modules.
This project aims at developing new big data software modules. Développement et déploiement web et distribué d'algorithmes d'analyse de données et d'intelligence artificielle pour les Big Data.
This project aims at developing image analysis reproducible benchmarking modules.
This projet aims at developing new AI modules for industrial quality control.








